library(shellfishrisks)
load_shellfish()
#> shellfishrisks is using Conda environment r-reticulate
batch <- "ctrl_demo_2"First, we’ll generate a control file with the default settings. This copies a .csv file named in this case “ctrl_file.csv” to your working directory. You could then open that file up and modify as you see fit.
ctrl_file_path <- generate_ctrl_file("ctrl_file")We’ll then run the shellfishrisk model using the settings in that control file. This will store the results in a folder called “results/{batch}”. A copy of the control file used to create that run will be stored in “results/{batch}” as well.
run_shellfishrisk_ctrlfile(batch = batch, reps = 1, ctrl_file_path = ctrl_file_path)And from there we can load and plot our results same as always.
results <- serve_shellfish(batches = batch) # read the results stored in .txt files into a list object
results$survival
#> # A tibble: 11 × 6
#> # Groups: batch, coreid, Rep [1]
#> batch coreid Rep age survival survivorship
#> <chr> <int> <int> <dbl> <dbl> <dbl>
#> 1 ctrl_demo_2 1 0 0 0.536 1
#> 2 ctrl_demo_2 1 0 1 0.393 0.536
#> 3 ctrl_demo_2 1 0 2 0.0429 0.211
#> 4 ctrl_demo_2 1 0 3 0.841 0.00903
#> 5 ctrl_demo_2 1 0 4 0.702 0.00759
#> 6 ctrl_demo_2 1 0 5 0.633 0.00533
#> 7 ctrl_demo_2 1 0 6 0.642 0.00338
#> 8 ctrl_demo_2 1 0 7 0.644 0.00217
#> 9 ctrl_demo_2 1 0 8 0.685 0.00140
#> 10 ctrl_demo_2 1 0 9 0.205 0.000957
#> 11 ctrl_demo_2 1 0 10 0 0.000196
plot_shellfish(results, type = "rvars")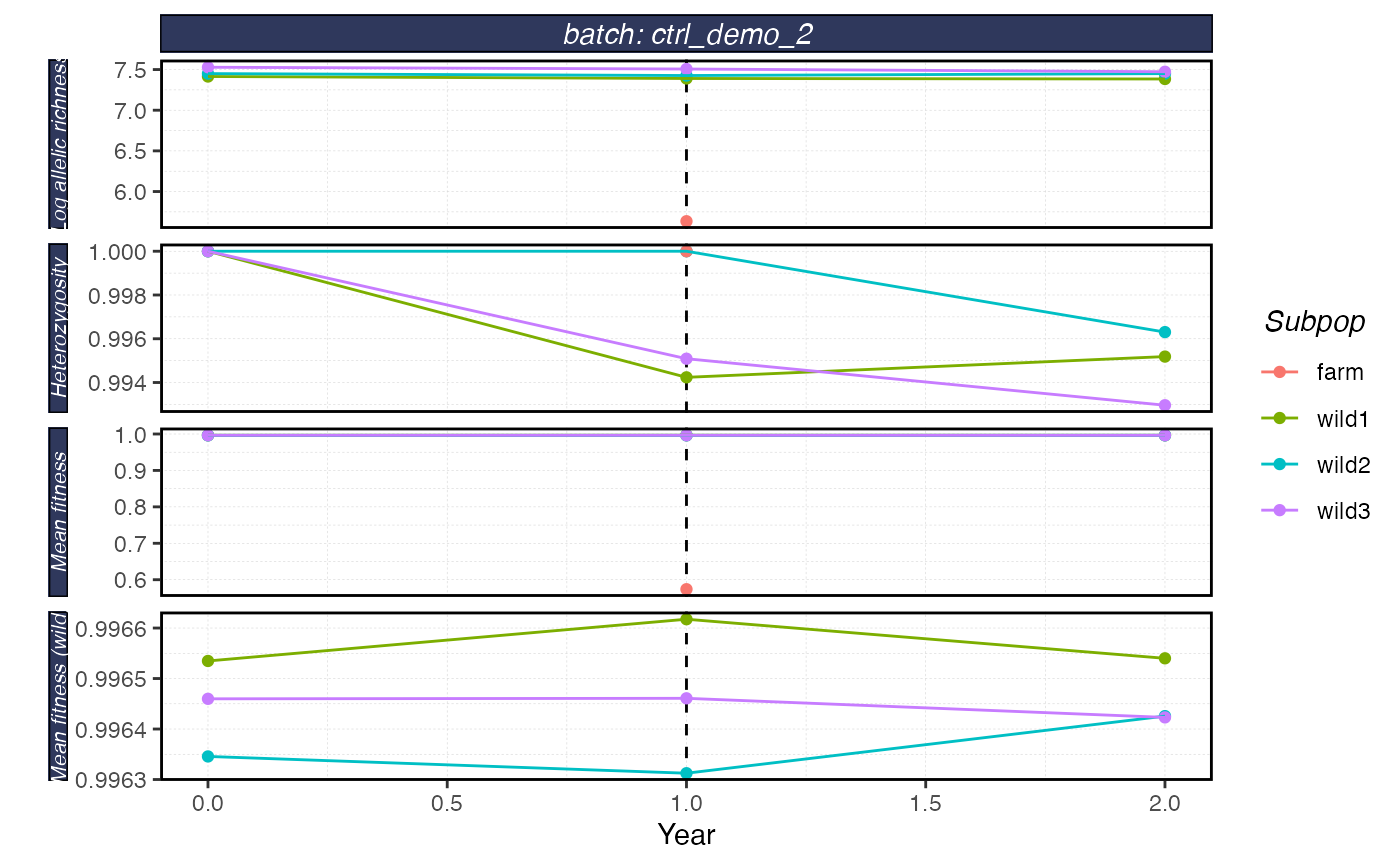
plot_shellfish(results, type = "fst")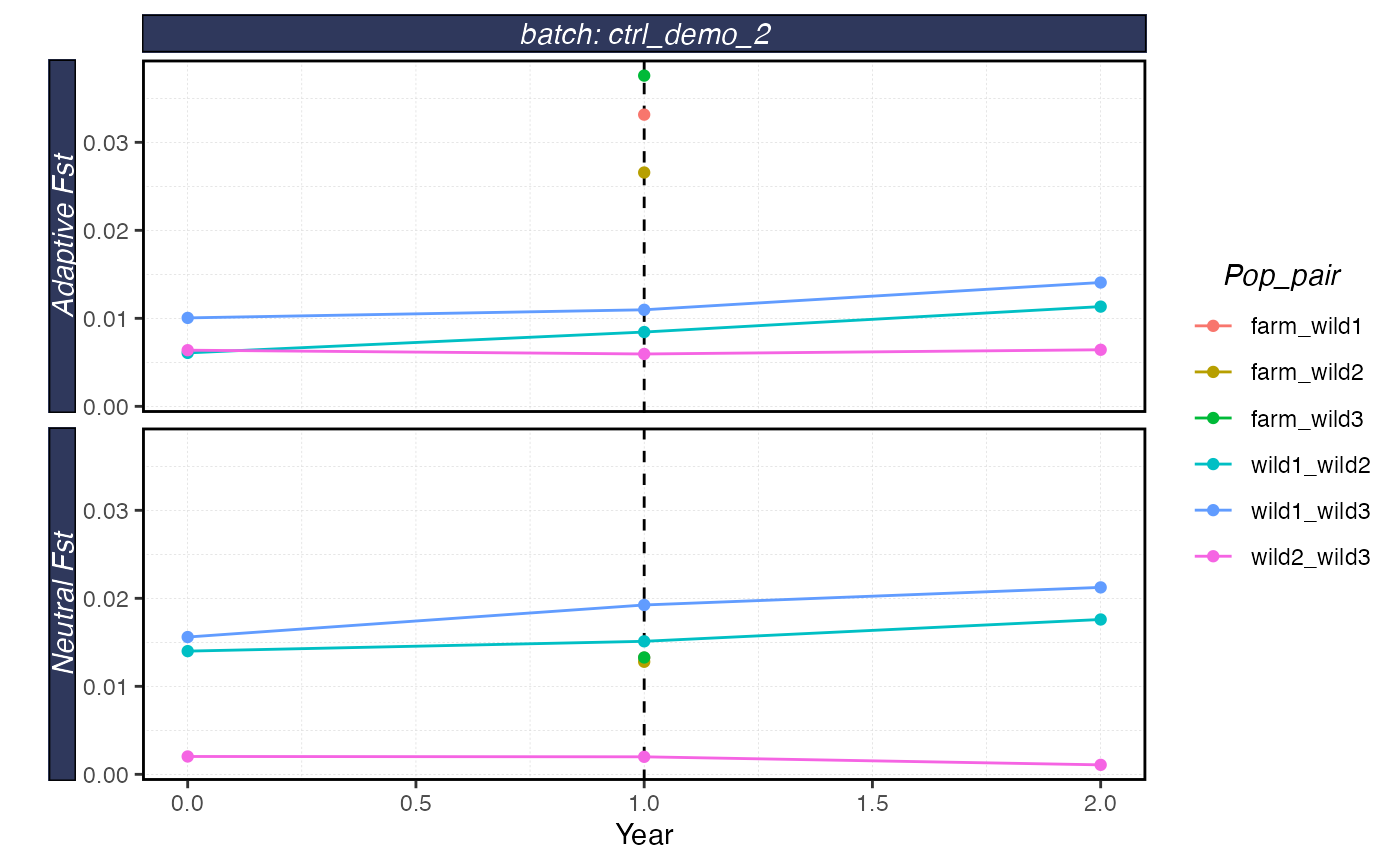
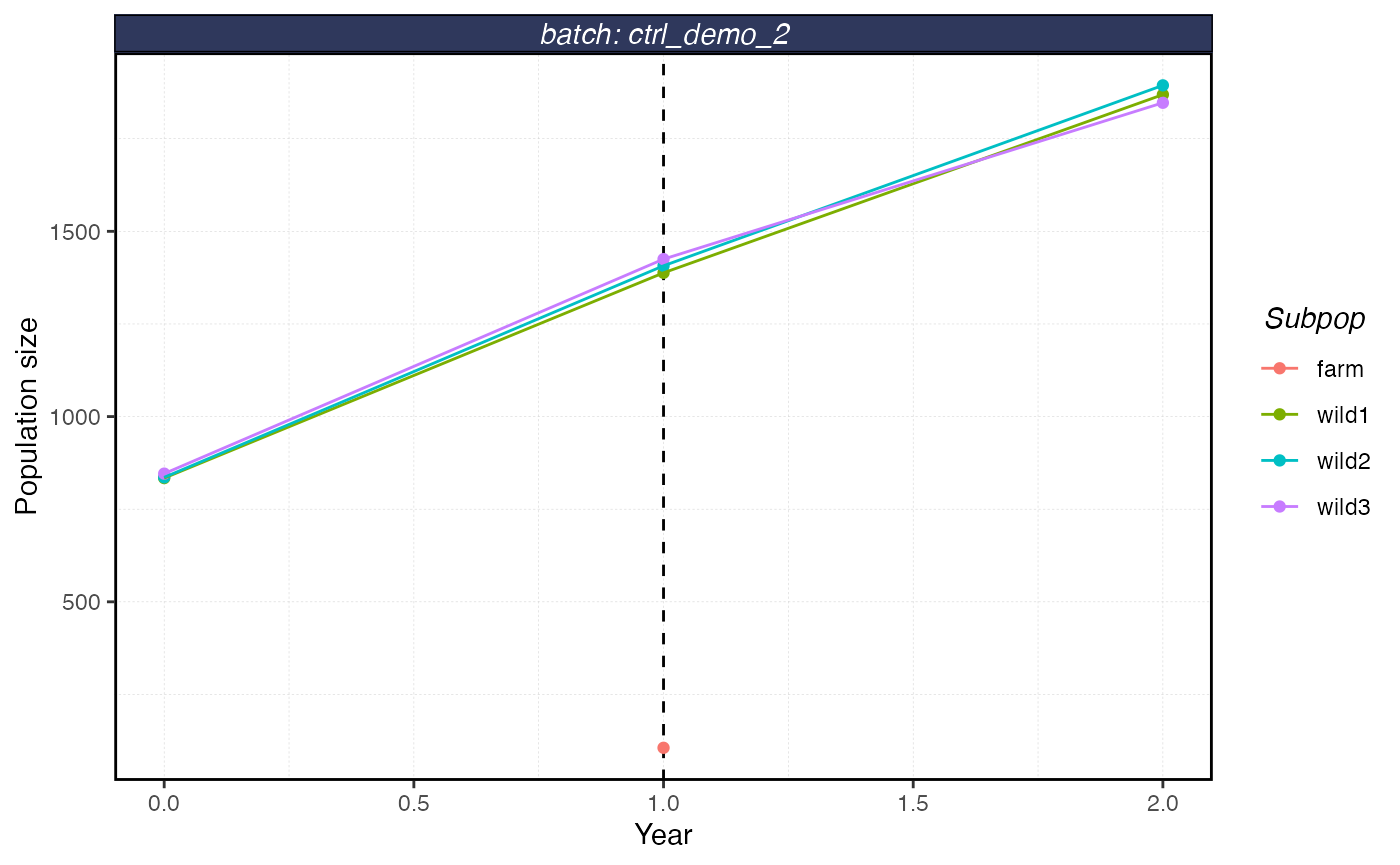
plot_shellfish(results, type = "popsize")